Transcriptomic Maps of Colorectal Liver Metastasis: Machine Learning of Gene Activation Patterns and Epigenetic Trajectories in Support of Precision Medicine
written by K. Rottmayer, H. Loeffler-Wirth, T. Gruenewald, Ilias. Doxiadis, C. Lehmann.
SIMPLY SUMMARY
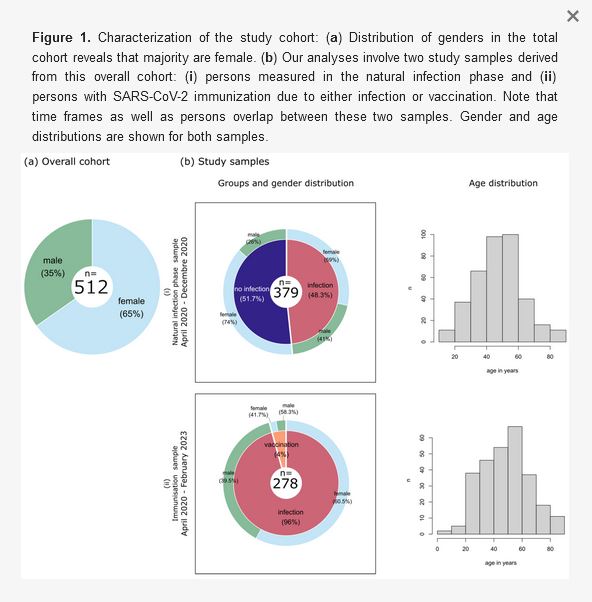
The immune response towards SARS-CoV-2 during the recent pandemic was analyzed retrospectively in a cohort of 512 individuals comprising 334 females and 178 males. In total, 737 sera were tested against several proteins of the virus and six seasonal coronaviruses. Differential reactivity was observed, and different HLA proteins were found to be associated with the antibody response towards SARS-CoV-2. This allowed us to identify low- and high-responder individuals. These results are indicative for an individual host immune response and may enable future individualized protective measures.
ABSTRACT
During the coronavirus pandemic, evidence is growing that the severity, susceptibility and host immune response to SARS-CoV-2 infection can be highly variable. Several influencing factors have been discussed. Here, we investigated the humoral immune response against SARS-CoV-2 spike, S1, S2, the RBD, nucleocapsid moieties and S1 of seasonal coronaviruses: hCoV-229E, hCoV-HKU1, hCoV-NL63 and hCoV-OC43, as well as MERS-CoV and SARS-CoV, in a cohort of 512 individuals. A bead-based multiplex assay allowed simultaneous testing for all the above antigens and the identification of different antibody patterns. Then, we correlated these patterns with 11 HLA loci. Regarding the seasonal coronaviruses, we found a moderate negative correlation between antibody levels against hCoV-229E, hCoV-HKU1 and hCoV-NL63 and the SARS-CoV-2 antigens. This could be an indication of the original immunological imprinting. High and low antibody response patterns were distinguishable, demonstrating the individuality of the humoral response towards the virus. An immunogenetical factor associated with a high antibody response (formation of ≥4 different antibodies) was the presence of HLA A*26:01, C*02:02 and DPB1*04:01 alleles, whereas the HLA alleles DRB3*01:01, DPB1*03:01 and DB1*10:01 were enriched in low responders. A better understanding of this variable immune response could enable more individualized protective measures.
to read the full article, click here: https://www.mdpi.com/2079-7737/12/10/1293?fbclid=IwAR11KgUqtr0W66DupwSRcCsLp099NRNLKGZSHcIqN6Nl_xIltQDr604Y4O4
