IMMUNOGENETIC PREDISPOSITION TO SARS-CoV-2 INFECTION
written by C. Lehmann, H. Loeffler-Wirth, V. Balz, J. Enczmann, R. Landgraf, N. Lakowa, T. Gruenewald, J. C. Fischer, I. Doxiadis.
Biology2023, 12(1), 37; https://doi.org/10.3390/biology12010037
SIMPLE SUMMARY
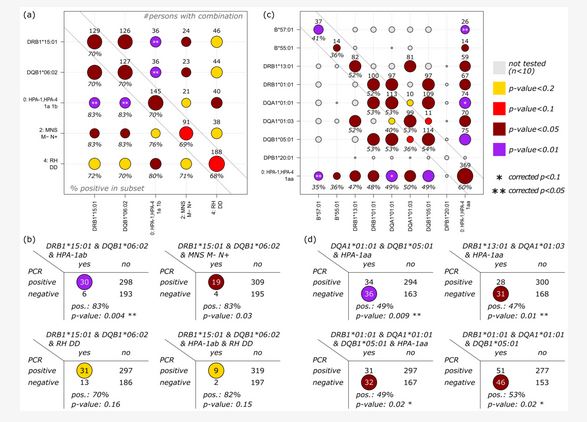
Since the beginning of the SARS-CoV-2 pandemic in 2020, numerous data with respect to the influence of immunogenetics on the predisposition to and severity of infection have been reported worldwide (PubMed; n = 228; 6 November 2022). Immunogenetics play a pivotal role in infection, vaccination, its failures, and/or vaccination breakthrough. Factors including the major histocompatibility complex and the common ABO blood group system have been discussed. Herein, we describe the association of HLA-A, B, C, DRB1, DRB345, DQA1, DQB1, DPA1, DPB1, and HLA-E, F, G, and H on the results of molecular detection of COVID-19, or, in some cases, on antibody detection upon first testing. Furthermore, we molecularly defined 22 blood group systems comprising 26 genes and 5 platelet antigen genes. Herein, 37% tested COVID-19 negative while 63% tested positive by PCR. Within the negative subjects, HLA-B*57:01, HLA-B*55:01, DRB1*13:01, and DRB1*01:01, were enriched, and in the positive group, homozygosity for DQA/DQB, DRB1*09:01, and DRB1*15:01 was observed. For HLA-DQA1, we observed an enrichment for DQA1*01:01, DQA1*02:01, and DQA1*01:03. For HLA-DQB1, we found that HLA-DQB1*06:02 was enriched in the positive group, while HLA-DQB1*05:01 and HLA-DQB1*06:03 were enriched in the negative group. The homozygous platelet antigen HPA-1a was significantly enriched in the negative group, contrasting with the HPA-1ab, which was enriched in the COVID-19 infected group.
ABSTRACT
Herein, we included 527 individuals from two Hospitals, Chemnitz and University-Hospital Leipzig. In total, 199 were negative for PCR and 328 were positive upon first admission. We used next generation sequencing for HLA-A, B, C, DRB1, DRB345, DQA1, DQB1, DPA1, and DPB1, and in some cases, HLA-E, F, G, and H. Furthermore, we molecularly defined 22 blood group systems comprising 26 genes and 5 platelet antigen genes. We observed a significant enrichment of homozygosity for DQA/DQB in the positive group. Within the negative subjects, HLA-B*57:01, HLA-B*55:01, DRB1*13:01, and DRB1*01:01 were enriched, and in the positive group, homozygosity for DQA/DQB, DRB1*09:01, and DRB1*15:01 was observed. DQA1*01:01, DQA1*02:01, and DQA1*01:03 were enriched in the negative group. HLA-DQB1*06:02 was enriched in the positive group, and HLA-DQB1*05:01 and HLA-DQB1*06:03 were enriched in the negative group. For the blood group systems MNS, RH, LE, FY, JK, YT, DO, and KN, enrichment was seen in both groups, depending on the antigen under observation. Homozygosity for D-positive RHD alleles, as well as the phenotypes M-N+ of the MNS blood group system and Yk(a-) of the KN system, were enriched in the positive group. All of these significances disappeared upon correction. Subjects who carried homozygous HPA-1a were more frequent in the negative group, contrasting with the finding that HPA-1ab was enriched in the positive group.
INTRODUCTION
The 2020–2022 COVID-19 (coronavirus) pandemic, caused by the SARS-CoV-2 virus, affected a large part of the population worldwide. This was especially true in Saxony, which showed the highest infections rates (46.8% in November 2022) in Germany [1]. Older people and individuals with previous illnesses, particularly, had reduced chances of survival. In daily life, pathogens induce immune responses in humans via two main pathways: the innate and the adaptive immune system, introduced by [2]. Herein, we concentrate on the later immune reactivity towards the SARS-CoV-2 virus [3]. The term immunogenetics describes a variety of genes involved in immunological responses against pathogens. These are inherited and generally polymorphic, i.e., have a great variability. The first Nobel Prize in the field of immunogenetics was awarded to Baruj Benacerraf, Jean Dausset, and George Davis Snell in 1980 for discovering genetically determined cellular surface structures, which control immunological reactions: “The Nobel Prize in Physiology or Medicine 1980” [4]. The genes encoded by the MHC (major histocompatibility complex) region, the HLAsystem (human leukocyte antigen), are expressed on the surface of nucleated cells and act as the counterpart of T and NK (natural killer) cell receptors, contributors to innate immunity. Upon infection, peptides of the intruder are presented at the cell membrane in the context of the MHC molecules: HLA-DR, DQ, and DP present peptides to the CD4 (cluster of differentiation) positive helper T cells, and HLA-A, B, and C, as well as the non-classical genes HLA-E and HLA-G, will present peptides to the CD8 positive cytotoxic T cells. HLA-F seems to present large peptides, but its precise function has not yet been elucidated [5]. The HLA nomenclature has been established through several workshops and accepted by the WHO nomenclature committee for factors of the HLA-System [6].
The MHC, as part of the immune system, plays a crucial role in the defense against pathogens [7]. Special variants of the MHC are associated with increased risk and complications of diseases, and have been found to be associated with viral infections [8,9]. For example, HIV is predominantly associated with MHC class I alleles, and Hepatitis B and Papilloma virus infections with MHC class II. In contrast, Dengue fever and Hepatitis C are associated with MHC class I and II alleles or allele groups [8]. An association between HLA-B27 and HLA-B57 was described for HIV [10], SARS-CoV [11], and SARS-CoV-2 [12]. In addition, a number of allelic variants of blood groups are also linked to infection susceptibility [13,14]. Herein, we searched for genetic factors within MHC and blood groups, which may determine susceptibility or resistance to SARS-CoV-2 infection. Genetic variants of the human SARS-CoV-2 receptors might be taken into consideration [15]. On the other hand, blood groups, which are, in general, multi-allelic and encoded on several regions of the human genome, were shown to be associated with viral infections [13]. We hypothesize that the previous observations of the propensity to develop immune recognition, and, hence, immune reaction, against SARS-CoV-2 depends on the allele status of HLA and blood group molecules in the infected individuals.
Several groups worldwide have reported associations with alleles/antigens of the HLA system. Besides single antigens, antigen groups or alleles were found to be associated with SARS-CoV-2 infection, severity, death, and survival [16,17,18,19,20,21,22,23]. However, the number of individuals tested was relatively small, and underlying diseases such as end-stage renal disease, autoimmunity, and immunosuppression might have affected the outcome. For the human platelet system (HPA), no data are yet available. For the common blood groups ABO and RHD, associations were observed, but the observations could not always be confirmed [14,17,18,24,25,26,27].
We analyzed, by next generation sequencing (NGS), the 11 classical loci for HLA (A, B, C, DRB1, DRB3, DRB4, DRB5, DQA1, DQB1, DPA1, and DPB1) and, for some of the individuals, the non-classical HLA genes, for a total of 26 red cell blood groups and 5 groups of the HPA system. Several of the alleles or allele groups were found to be primarily associated with the SARS-CoV-2 infection, with a p value of <0.1.
…
to read the full article at MDPI, please click here: https://www.mdpi.com/2079-7737/12/1/37#
