THE GENOMIC AND TRANSCRIPTIONAL LANDSCAPE OF PRIMARY CENTRAL NERVOUS SYSTEM LYMPHOMA
written by Josefine Radke,Naveed Ishaque,Randi Koll,Zuguang Gu,Elisa Schumann,Lina Sieverling,Sebastian Uhrig,Daniel Hübschmann,Umut H. Toprak,Cristina López,Xavier Pastor Hostench,Simone Borgoni,Dilafruz Juraeva,Fabienne Pritsch,Nagarajan Paramasivam,Gnana Prakash Balasubramanian,Matthias Schlesner,Shashwat Sahay,Marc Weniger,Debora Pehl,Helena Radbruch,Anja Osterloh,Agnieszka Korfel,Martin Misch,Julia Onken,Katharina Faust,Peter Vajkoczy,Dag Moskopp,Yawen Wang,Andreas Jödicke,Lorenz Trümper,Ioannis Anagnostopoulos,Dido Lenze,Ralf Küppers,Michael Hummel,Clemens A. Schmitt,Otmar D. Wiestler,Stephan Wolf,Andreas Unterberg,Roland Eils,Christel Herold-Mende,Benedikt Brors,ICGC MMML-Seq Consortium,Reiner Siebert,Stefan Wiemann &Frank L. Heppner
ABSTRACT
Primary lymphomas of the central nervous system (PCNSL) are mainly diffuse large B-cell lymphomas (DLBCLs) confined to the central nervous system (CNS). Molecular drivers of PCNSL have not been fully elucidated. Here, we profile and compare the whole-genome and transcriptome landscape of 51 CNS lymphomas (CNSL) to 39 follicular lymphoma and 36 DLBCL cases outside the CNS. We find recurrent mutations in JAK-STAT, NFkB, and B-cell receptor signaling pathways, including hallmark mutations in MYD88 L265P (67%) and CD79B (63%), and CDKN2A deletions (83%). PCNSLs exhibit significantly more focal deletions of HLA-D (6p21) locus as a potential mechanism of immune evasion. Mutational signatures correlating with DNA replication and mitosis are significantly enriched in PCNSL. TERT gene expression is significantly higher in PCNSL compared to activated B-cell (ABC)-DLBCL. Transcriptome analysis clearly distinguishes PCNSL and systemic DLBCL into distinct molecular subtypes. Epstein-Barr virus (EBV)+ CNSL cases lack recurrent mutational hotspots apart from IG and HLA-DRB loci. We show that PCNSL can be clearly distinguished from DLBCL, having distinct expression profiles, IG expression and translocation patterns, as well as specific combinations of genetic alterations.
INTRODUCTION
Central nervous system (CNS) lymphomas are predominantly aggressive neoplasms involving brain, meninges, spinal cord, and eyes1,2. Two clinical subtypes of CNSL can be distinguished: primary central nervous system lymphoma (PCNSL), which is confined to the CNS; and secondary central nervous system lymphoma (SCNSL) presenting initially with systemic, non-CNS or synchronous systemic and CNS involvement. The term SCNSL comprises all systemic lymphomas that spread to the CNS and its presentation, tropism, outcome and therapeutic options differ from PCNSL3,4. Typically, SCNSL are classified as diffuse large B-cell lymphoma (DLBCL), while other types such as follicular lymphoma (FL), T-cell lymphoma or Hodgkin lymphoma are extremely rare5,6.
PCNSL incidence is increased in immunocompromised patients, in which the tumor cells are typically Epstein-Barr virus (EBV)-positive1,7,8. In contrast, PCNSL in immunocompetent patients is typically EBV-negative. The mechanisms leading to the observed exclusive topographical restriction of PCNSL to the CNS are not fully elucidated9. PCNSL is classified as DLBCL in the vast majority of cases (approx. 90%) which immunohistochemically most often show a non-germinal center B-cell-like (non-GCB) immunophenotype1,10,11 according to the Hans classification12. The tumor cells express pan B-cell markers (CD19, CD20, and CD79a), the germinal center (GC)-associated molecule BCL613, and the post-GC-associated marker MUM1/IRF414. By gene expression profiling, the tumor cells are most closely related to late germinal center (exit) B-cells15. Pathomechanistic genomic alterations involving Toll-like- and B-cell receptor (TLR, BCR) signaling pathways are identified in previous studies revealing a very high frequency of somatic nonsynonymous mutations in genes such as MYD88, CARD11, and CD79B16,17,18,19,20. Additionally, often homozygous HLA class II21,22 and CDKN2A loss, recurrent BCL6 translocations23,24 and structural variants at chromosome band 9p24.1 (affecting CD274/PD-L1 and PDCD1LG2/PD-L2)25 as well as TBL1XR1 variants26 are repeatedly described in PCNSL27,28. These mutational patterns suggest PCNSL to be genetically similar to recently described “MCD”, “C5” or “MYD88-like” subtypes for which a derivation from long-lived memory B-cells is proposed29,30,31,32,33,34,35.
The outcome of PCNSL, even in immunocompetent hosts, is poor compared to most primary systemic DLBCL36, though probably not worse than that of DLBCL of the MCD/C5 group in general30. High-dose methotrexate (MTX) remains the commonly administered therapy but the use of rituximab (monoclonal anti-CD20 antibody) is shown to be effective37,38. However, reports on rituximab efficiency in PCNSL are conflicting39,40,41,42. Genomic studies suggest that lymphoma cell proliferation and survival are driven at least in part, by deregulated TLR, BCR, JAK-STAT, and NFκB signaling pathways inducing constitutive NFκB activation43,44,45. Therefore, inhibitors up- and downstream of NFκB such as ibrutinib, known to inhibit Bruton’s tyrosine kinase (BTK) as critical mediator of BCR signaling, and lenalidomide which is shown to have indirect effects on tumor immunity are applied and seem to be effective therapeutic alternatives in PCNSL46,47,48,49,50,51. PD-L1/2 blockade is discussed as another therapeutic option52.
Despite all progress in the molecular characterization of PCNSL in the last decades, our understanding of the genetic and transcriptional alterations of PCNSL is by far not comprehensive. The few previous next generation sequencing (NGS) studies of PCNSL are limited to target enrichment only of exons, or whole-genome analysis of very few samples44,53,54,55,56,57,58. Therefore, pathogenic mechanisms other than coding variants, such as non-coding and regulatory changes, structural variants or mutational mechanisms related to the genome-wide distribution of somatic hypermutation (SHM) are not fully elucidated in PCNSL. Unbiased omics profiling, such as whole-genome sequencing (WGS) studies integrated with transcriptome sequencing, are currently the methods of choice to illuminate the role of non-coding mutations59,60. In addition, these approaches can unravel various molecular mechanisms deregulating driver genes in PCNSL, which are necessary for diagnosis, risk stratification, and treatment in the era of precision and targeted therapies.
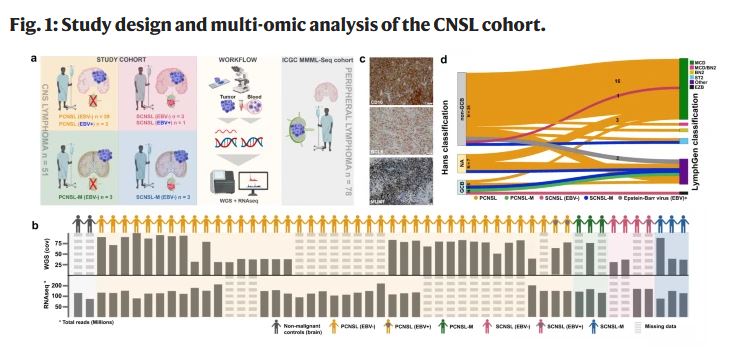
In this work, we perform whole-genome and transcriptome sequencing in 51 B-cell lymphomas presenting in the CNS, including 42 PCNSL samples from immunocompetent patients, to comprehensively describe the mutational and transcriptional landscape of PCNSL.
to read the full article, click here: https://www.nature.com/articles/s41467-022-30050-y
