written by Ashekyan, O.; Shahbazyan, N.; Bareghamyan, Y.; Kudryavzeva, A.; Mandel, D.; Schmidt, M., . . . Binder, H. (Armenian Bioinformatics Institute, 3/6 Nelson Stepanyan Str., Yerevan 0062, Armenia in cooperation with IZBI, Interdisciplinary Centre for Bioinformatics, Universität Leipzig, Härtelstr. 16–18, 04107 Leipzig, Germany and Agenus Inc., 3 Forbes Road, Lexington, MA 7305, USA and Institute of Molecular Biology of the National Academy of Sciences of the Republic of Armenia, 7 Has-Ratyan Str., Yerevan 0014, Armenia)
SIMPLE SUMMARY
Liver metastasis is a significant factor contributing to mortality associated with colorectal cancer. Establishing the biological mechanisms of metastasis is crucial for refining diagnostics and identifying therapeutic windows for interventions. Currently, little is known of the processes that govern the development of liver metastases, the role of the tumor microenvironment, the role of epigenetics, and potential treatment-induced shaping effects. Machine learning-based bioinformatics has provided an important methodical option to decipher fine-granular details of the transcriptomic landscape of tumor heterogeneity and the underlying molecular mechanisms. Our molecular portrayal method has potential implications for treatment decisions, which may require personalized diagnostics.
ABSTRACT
The molecular mechanisms of the liver metastasis of colorectal cancer (CRLM) remain poorly understood. Here, we applied machine learning and bioinformatics trajectory inference to analyze a gene expression dataset of CRLM. We studied the co-regulation patterns at the gene level, the potential paths of tumor development, their functional context, and their prognostic relevance. Our analysis confirmed the subtyping of five liver metastasis subtypes (LMS). We provide gene-marker signatures for each LMS, and a comprehensive functional characterization that considers both the hallmarks of cancer and the tumor microenvironment. The ordering of CRLMs along a pseudotime-tree revealed a continuous shift in expression programs, suggesting a developmental relationship between the subtypes. Notably, trajectory inference and personalized analysis discovered a range of epigenetic states that shape and guide metastasis progression. By constructing prognostic maps that divided the expression landscape into regions associated with favorable and unfavorable prognoses, we derived a prognostic expression score. This was associated with critical processes such as epithelial–mesenchymal transition, treatment resistance, and immune evasion. These factors were associated with responses to neoadjuvant treatment and the formation of an immuno-suppressive, mesenchymal state. Our machine learning-based molecular profiling provides an in-depth characterization of CRLM heterogeneity with possible implications for treatment and personalized diagnostics.
INTRODUCTION
Liver metastasis, linked with poor prognosis, is a common occurrence in various cancers, including colorectal cancer (CRC), pancreatic cancer, breast cancer, melanoma, and lung cancer. In CRC, the liver is the primary site of metastasis owing to the anatomical and vascular connections between the colorectal regions and the liver [1]. Such metastases pose a significant challenge for clinical intervention and represent a major cause of CRC-related mortality. However, the molecular mechanisms that govern the molecular heterogeneity and tumor development in liver metastasis remain poorly understood.
The tumor microenvironment (TME) significantly impacts the pathophysiology of cancer cells metastasizing to the liver. It includes liver sinusoidal endothelial cells, Kupffer cells, hepatic stellate cells, and parenchymal hepatocytes, as well as infiltrating stromal and immune cells [2]. Interactions with the TME facilitate cancer cells to overcome the tumor stroma, settle, and to colonize. Interestingly, colorectal cancer liver metastases (CRLM) show high genetic concordance in key lesions, mutations, and copy number variations (CNV) with primary CRC suggesting the liver microenvironment has a limited influence on the mutation pattern of CRLM cells, which remain largely genetically primed by their primary CRC origin [3]. This correlation of molecular characteristics between primary and metastatic tumors is further supported by the functional and transcriptional studies on CRLM [3,4,5,6,7]. However, genetic lesions and adaptive interactions with the TME, while necessary, are not sufficient for cancer initiation and progression. As a third factor, epigenetic regulation, including chromatin remodeling associated and driven by a large set of histone- and DNA-modifying mechanisms, are essential for cancer clones to acquire the plasticity necessary for adaptive cell fate changes towards evolutionary fitness in an epigenetic landscape [8,9]. Currently, little is known of the evolutionary processes that govern CRLM, the role of epigenetics, and potential treatment-induced shaping effects. Establishing the biological mechanisms of metastasis is crucial for refining diagnostics and identifying therapeutic windows for interventions.
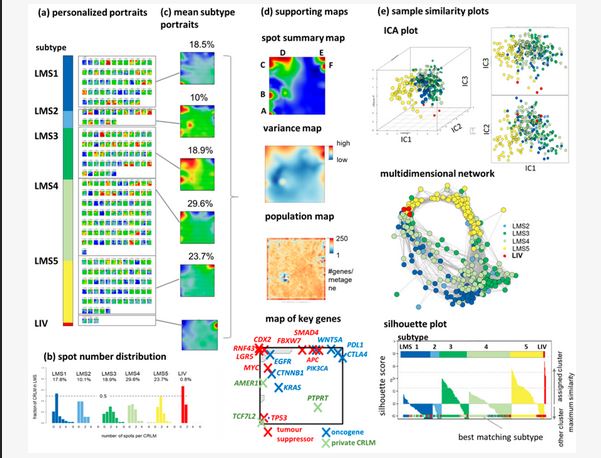
Molecular subtyping has emerged as an important concept to decipher cancer heterogeneity in both primary and metastatic tumors [10]. While correlations between the molecular subtypes of primary CRC, metastatic propensity, and responses to therapy have been noted [6], it is not clear to what degree these CRC characteristics are maintained after distant spread. Recently, Moosavi et al. developed a metastasis-oriented subtyping framework through the transcriptomic analysis of patients with CRLMs [5]. Their de novo liver metastases subtypes (LMS) recapitulated epithelial-like and mesenchymal-like tumors, with the latter showing a strong immune and stromal component.
In this study, we leveraged machine learning-based bioinformatics to analyze the transcriptomes of 283 CRLM [5], to characterize the molecular landscape with single-tumor resolution, and to deduce cues for CRLM development under neoadjuvant treatment. We further evaluated and extended the LMS framework, and identified subtype-specific gene signatures, explored their functional relevance to cancer progression, and suggest possible developmental paths under epigenetic control using pseudotime inference through in-depth transcriptomic analysis. Our approach utilizes a high-resolution molecular cartography and portrayal method previously applied to various cancer types [11,12,13,14], treatment resistance [15], and modes of epigenetic regulation [16,17]. In parallel, we have established, and made available, an interactive web platform for more detailed insights into our analyses. Through the personalized portrayal of gene expression data and the creation of prognostic maps for CRLM, we open new avenues for personalized diagnostics and treatment decision-making.
. . .
CONCLUSIONS
Machine learning using an omics portrayal of CRLM provides a comprehensive and detailed understanding of the molecular heterogeneity underlying this complex disease. Our study revealed a shift from treatment-sensitive to treatment-resistant tumors that are guided by genetic, epigenetic, and microenvironmental factors. Our findings encourage further studies to better understand the micro-spatial heterogeneity of the CRLM, the underlying epigenetic mechanisms, the changing cellular communities in the TME interacting with the tumor cells, and also possible genetic determinants. Understanding this heterogeneity is crucial for tailoring effective treatment strategies, where precision medicine is one promising approach. It must involve the genomic profiling of individual tumors in follow-up settings to identify specific alterations and vulnerabilities in space and time to tackle the diverse manifestations of CRLM and provide better control of disease progression. This information can guide the selection of targeted therapies, immunotherapies, or combined treatments.
>>> to read the full article, go here: https://www.mdpi.com/2072-6694/15/15/3835
