UNVEILING ISO- & ANISO-HYDRIC DISPARITIES IN GRAPEVINE – A Reanalysis by Transcriptome Portrayal Machine Learning
written by Tomas Konecny, Armine Asatryan, Maria Nikoghosyan, PD Hans Binder
ABSTRACT
Mechanisms underlying grapevine responses to water(-deficient) stress (WS) are crucial for viticulture amid escalating climate change challenges. Reanalysis of previous transcriptome data uncovered disparities among isohydric and anisohydric grapevine cultivars in managing water scarcity. By using a self-organizing map (SOM) transcriptome portrayal, we elucidate specific gene expression trajectories, shedding light on the dynamic interplay of transcriptional programs as stress duration progresses. Functional annotation reveals key pathways involved in drought response, pinpointing potential targets for enhancing drought resilience in grapevine cultivation. Our results indicate distinct gene expression responses, with the isohydric cultivar favoring plant growth and possibly stilbenoid synthesis, while the anisohydric cultivar engages more in stress response and water management mechanisms. Notably, prolonged WS leads to converging stress responses in both cultivars, particularly through the activation of chaperones for stress mitigation. These findings underscore the importance of understanding cultivar-specific WS responses to develop sustainable viticultural strategies in the face of changing climate.
INTRODUCTION
Vitis vinifera, being a cornerstone in the global wine industry, faces unprecedented challenges in the era of climate change. Among these challenges, drought stands out as one of the pivotal factors causing water(-deficient) stress (WS), influencing grape ripening and overall fruit quality. As global temperatures rise and precipitation patterns become increasingly unpredictable, the study of WS in Vitis vinifera has become paramount for ensuring the resilience of vineyards and the sustainability of wine production.
Current climate change scenarios have brought about shifts in weather patterns, leading to prolonged periods of water scarcity and extreme climatic events. These changes pose a significant threat to viticulture, impacting grapevine physiology, grape quality, and wine production. WS, characterized by a deficiency in water availability relative to the plant’s demand, triggers a cascade of physiological responses in Vitis vinifera, affecting its growth, photosynthesis, and metabolic processes [1,2,3,4]. Notably, grapevines have evolved two distinct mechanisms, isohydric and anisohydric, to navigate the complex interplay between water availability and demand [5,6,7,8,9]. Isohydric cultivars regulate water potential by actively adjusting stomatal conductance, conserving water during limited availability, and prioritizing water status regulation over continuous carbon assimilation. Conversely, anisohydric cultivars maintain sustained carbon assimilation even under declining water availability, with a more permissive control over stomatal conductance [10,11,12]. While promoting growth and productivity, this strategy exposes grapevines to higher risks of hydraulic failure under severe water deficit situations [9,13,14].
The investigation of these isohydric and anisohydric mechanisms gains significance in deciphering the adaptive strategies of Vitis vinifera to varying water conditions [15,16,17]. The importance of such research is heightened by the need to develop resilient grape cultivars or drought-resistant rootstocks and implement sustainable viticultural practices in response to the changing climate.
Even though both isohydric and anisohydric mechanisms in different grapevine cultivars are well-known and studied at the transcriptomics level [15], current advancements in the field of artificial intelligence (AI) and machine learning (ML) and their applications in analyzing molecular biology data offer exciting possibilities. AI- and ML-powered analyses, particularly through techniques like self-organizing maps (SOM), have the potential to unlock a deeper understanding of the underlying mechanisms and biological pathways that occur at specific time points during stress treatments relevant to viticulture [18]. SOM is a type of artificial neural network that can visualize high-dimensional data like gene expression profiles. By analyzing transcriptomic data using SOM, hidden patterns and relationships between genes can be identified and visualized, enhancing understanding of the underlying mechanisms governing grapevine responses to abiotic stress conditions caused, for instance, by cold temperatures or by water limitation.
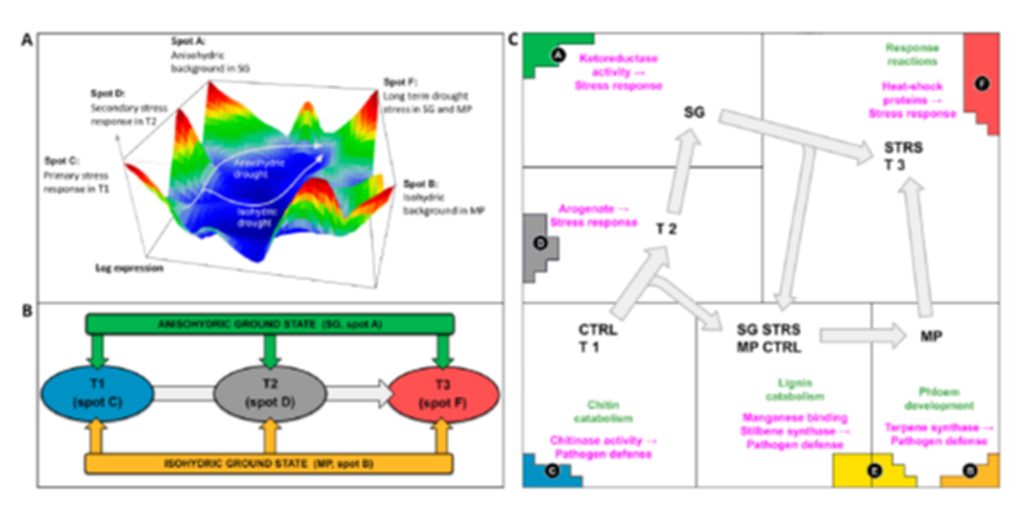
A previous study [15] delved into how grapevine transcriptomes respond to WS at the molecular level. The studied isohydric cultivar (‘Montepulciano’) exhibited a faster transcriptome response after WS imposition, with rapid modulation of genes related to abscisic acid (ABA), a stress response hormone, and quicker expression of heat-shock protein (HSP) genes. On the other hand, an anisohydric cultivar (‘Sangiovese’) displayed early and robust induction of reactive oxygen species (ROS)-scavenging enzymes, molecular chaperones, and abiotic-stress-related genes in response to deficiency of water. While this research provides valuable insights, it is important to recognize that the genes identified by significance analysis of microarrays (SAM) represent a portion of the grapevine’s response to WS [19]. Here, we apply SOM omics portrayal to these data to analyze the transcriptomic response of isohydric or anisohydric grapevines under water deficit conditions. SOM transcriptomics portrayal was developed to extract and interpret complex patterns from multidimensional transcriptomic data [20], and it has been applied recently to cold stress acting on Vitis vinifera and, particularly, to describe the transcriptomics trajectories of the grapevine plant under changing environmental conditions [18]. Our aim is to compare trajectories associated with drought response in the isohydric and anisohydric grapevine categories. We show that isohydric MP initially maintains a muted transcriptional response, prioritizing transcripts for normal growth (phloem development) before potentially shifting towards activation of stress response processes for survival. Conversely, anisohydric SG displays a rapid transcriptional response, initially focusing on stress alleviation (ketoreductase, arogenate) and water management (lignin catabolism, chitinase activity) before upregulating thiamin biosynthesis, a pathway potentially linked to broader abiotic stress tolerance. Despite their distinct genetic backgrounds related to water-use efficiency, both cultivars eventually converge toward shared responses under prolonged drought conditions.
to read the full article, click here: https://www.mdpi.com/2223-7747/13/17/2501
