CLASSIFYING GERMINAL CENTER DERIVED LYMPHOMAS -NAVIGATE A COMPLEX TRANSCRIPTIONAL COMPLEX LANDSCAPE
written by Henry Loeffler-Wirth, Markus Kreuz, Maria Schmidt, German Ott, Reiner Siebert , Hans Binder
SIMPLE SUMMARY
Germinal center-derived B-cell lymphomas constitute a very heterogeneous group of neoplasms with diverse clinical presentations, prognoses, and responses to therapy. They divide into a series of subtypes, such as Diffuse Large B-cell lymphomas (DLBCL), Burkitt Lymphomas (BL), Follicular lymphomas (FL), and further, into several subclasses and rarer subtypes of finer granularity, which makes them one of the most heterogeneous cancer entities. Molecular classification schemes, first of all, derived from whole transcriptome gene expression data largely improved subtyping and functional understanding. Based on a whole transcriptome landscape of B-cell lymphoma, we show that one major caveat in the task of classification is represented by the rather fuzzy distribution of individual tumors without clear-cut borderlines between most of the subtypes, preventing their unambiguous association with clear-cut entities. This landscape is governed by the germinal center (GC) reaction and relates different subtypes to different states along the reaction path. We discuss the relatedness between the expression landscape and classifier signatures and their functional cell of origin background. This view helps to stratify lymphomas in terms of modular building blocks of signature genes and to interpret rarer subclasses in the context of larger ones.
ABSTRACT
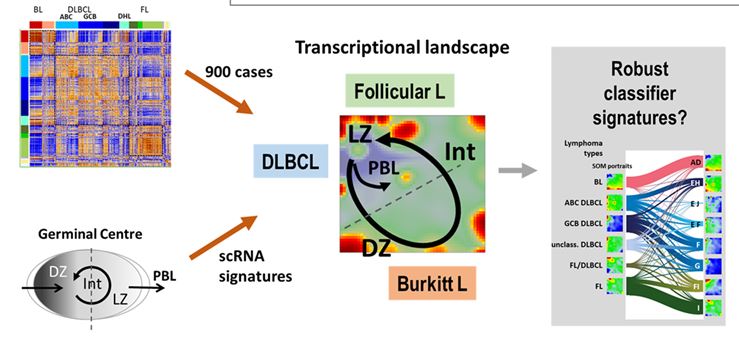
Classification of lymphoid neoplasms is based mainly on histologic, immunologic, and (rarer) genetic features. It has been supplemented by gene expression profiling (GEP) in the last decade. Despite the considerable success, particularly in associating lymphoma subtypes with specific transcriptional programs and classifier signatures of up- or downregulated genes, competing molecular classifiers were often proposed in the literature by different groups for the same classification tasks to distinguish, e.g., BL versus DLBCL or different DLBCL subtypes. Moreover, rarer sub-entities such as MYC and BCL2 “double hit lymphomas” (DHL), IRF4-rearranged large cell lymphoma (IRF4-LCL), and Burkitt-like lymphomas with 11q aberration pattern (mnBLL-11q) attracted interest while their relatedness regarding the major classes is still unclear in many respects. We explored the transcriptional landscape of 873 lymphomas referring to a wide spectrum of subtypes by applying self-organizing maps (SOM) machine learning. The landscape reveals a continuum of transcriptional states activated in the different subtypes without clear-cut borderlines between them and preventing their unambiguous classification. These states show striking parallels with single cell gene expression of the active germinal center (GC), which is characterized by the cyclic progression of B-cells. The expression patterns along the GC trajectory are discriminative for distinguishing different lymphoma subtypes. We show that the rare subtypes take intermediate positions between BL, DLBCL, and FL as considered by the 5th edition of the WHO classification of haemato-lymphoid tumors in 2022. Classifier gene signatures extracted from these states as modules of coregulated genes are competitive with literature classifiers. They provide functional-defined classifiers with the option of consenting redundant classifiers from the literature. We discuss alternative classification schemes of different granularity and functional impact as possible avenues toward personalization and improved diagnostics of GC-derived lymphomas.
…
CONCLUSIONS
The whole-transcriptome SOM landscape of GC-derived lymphomas in combination with portrayal of the individual tumors offers novel perspectives for consenting existing redundant classifier signatures, for their better functional understanding in the light of the GC reaction and for increasing granularity of classification towards personalized diagnostics. Consent between different gene expression signatures as well as their extension towards existing novel classification schemes such as PAT grouping, hallmark types, LME groups and single cell-derived COO classes, are needed to combine their different functional perspectives into a common view on the molecular phenotypes of lymphoma. The spot-modules of co-regulated signature genes represent “mountain peaks” used as classification landmarks in an otherwise continuous transcriptomic landscape of GC-derived lymphomas, which leaves classification uncertain to some residual degree. Still existing inconsistencies between genetic, transcriptomic and microenvironmental classifiers need further research with larger tumor cohorts to better resolve rare classes. Improved experimental techniques such as single-cell omics measurements on the data side and probabilistic classification schemes and integrative multi-omics bioinformatics in combination with machine learning on the analytics side are the next milestones along the avenue towards further improved, classification of lymphomas.
to read the full article, click here: https://www.mdpi.com/2072-6694/14/14/3434/htm
