HIGHLIGHT DATA SETS
ALL AVAILABLE DATA SETS
| Blood (healthy cohort) | Bulk Transcriptome | start browsing |
| Blood (CAR-T cell therapy; Sheih et al.) | Single-cell Transcriptome | start browsing |
| Blood (CAR-T cell therapy; Deng et al.) | Single-cell Transcriptome | start browsing |
| Colorectal cancer (tumor samples) | Bulk Transcriptome | start browsing |
| Colorectal cancer (tumor samples) | Bulk Transcriptome | start browsing |
| Colorectal cancer | Spatial Transcriptomics | start browsing |
| Colorectal cancer | Spatial Transcriptomics | start browsing |
| Duodenum (coeliac disease) | Bulk Transcriptome | start browsing |
| Fibroblasts | Single-cell Transcriptome | start browsing |
| Liver (metastases) | Bulk Transcriptome | start browsing |
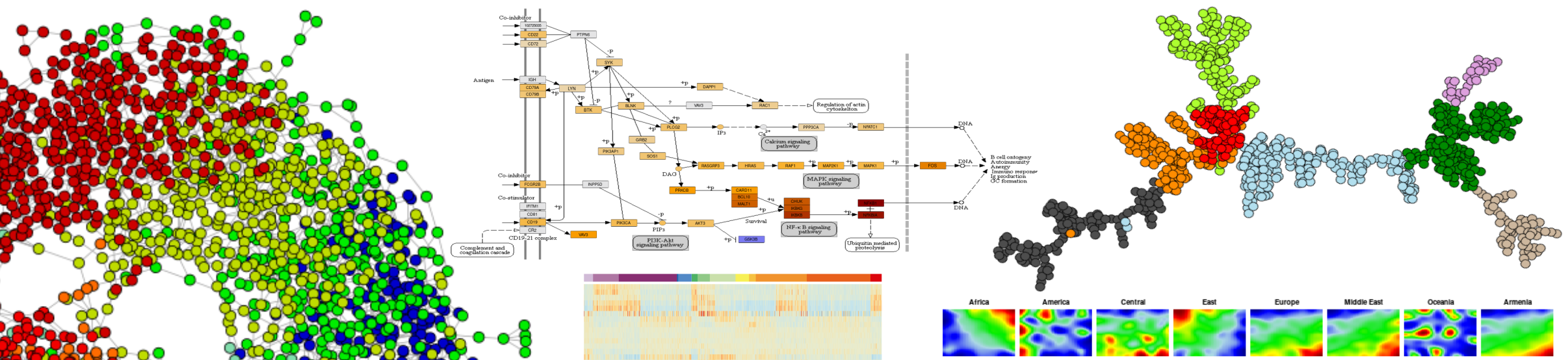
| Lymphoma | Bulk Transcriptome | start browsing |
| Glioma (low grade) | Bulk Transcriptome | start browsing |
| Glioma (low grade) | Methylome | start browsing |
| Glioma (grade 1-4) | Bulk Transcriptome | start browsing |
| Meibomian gland | Spatial Transcriptomics | start browsing |
| Melanoma (plus Naevi) | Bulk Transcriptome | start browsing |
| Melanoma (plus Naevi) | Single-cell Transcriptome | start browsing |
| Melanoma | Spatial Transcriptomics | start browsing |
| Pneumonia | Bulk Transcriptome | start browsing |
| Sebaceous gland | Spatial Transcriptomics | start browsing |
| Brain (healthy mouse) | Spatial Transcriptomics | start browsing |
| Brain (Thy1-wt & -ko, mouse) | Single-cell Transcriptomics | start browsing |
| Human genome atlas | Genome (SNPs) | start browsing |
| SARS-Cov2 virus atlas | GEnome (SNVs) | start browsing |
